A consensus blood transcriptomic framework for sepsis
Scicluna, B.P., Cano-Gamez, K., Burnham, K.L. et al.
Nat Med (2025). https://doi.org/10.1038/s41591-025-03964-5
Abstract
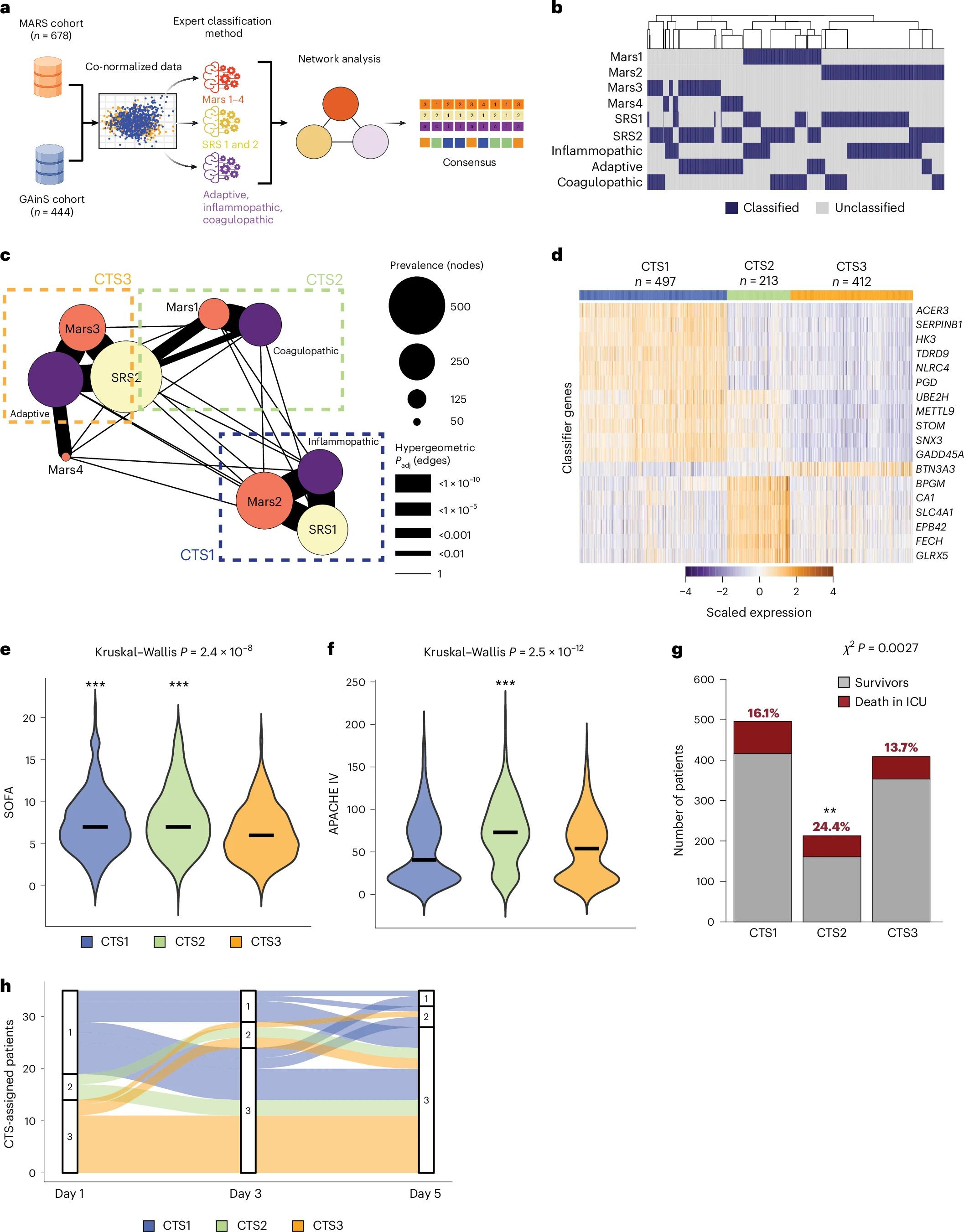
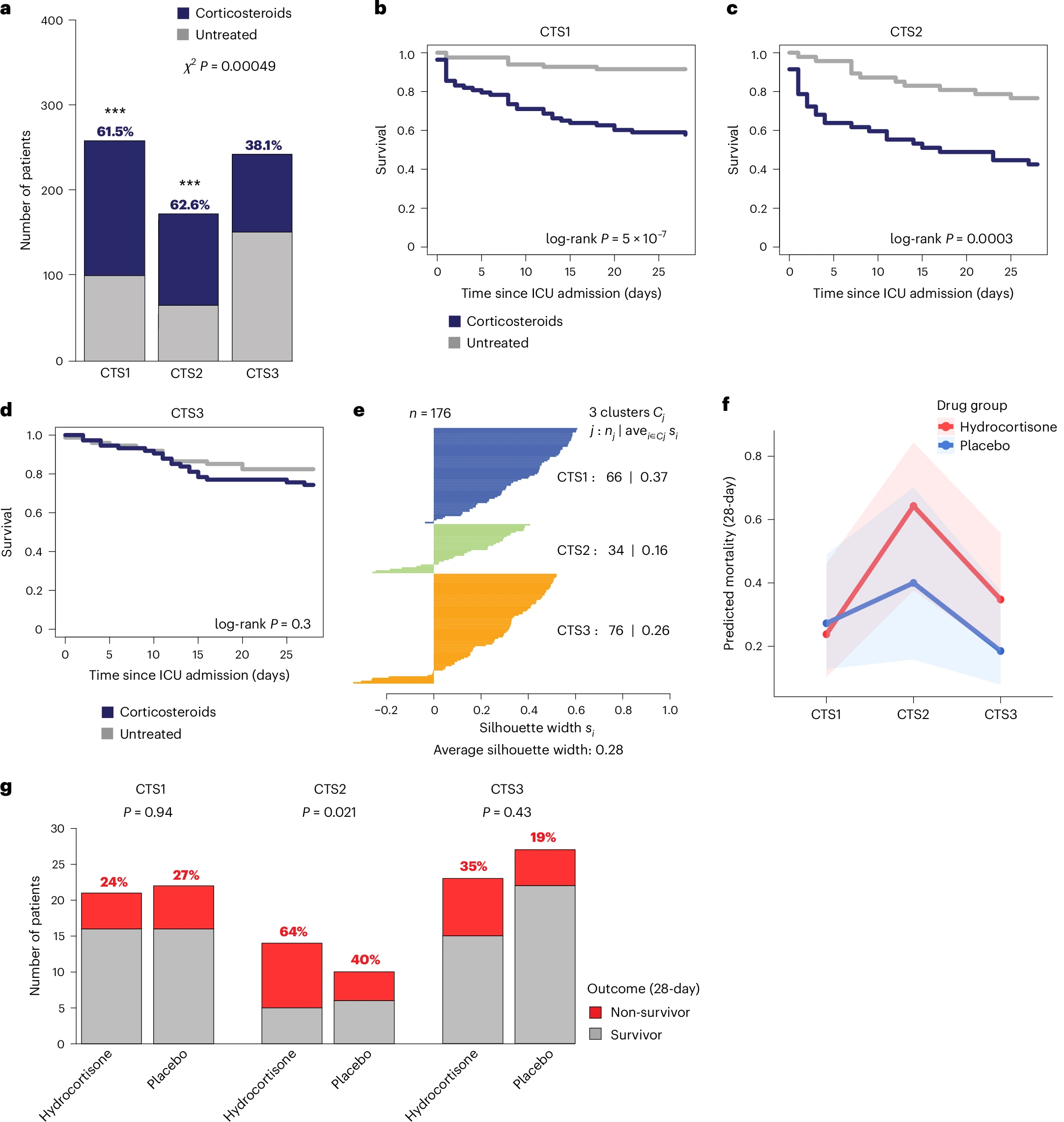
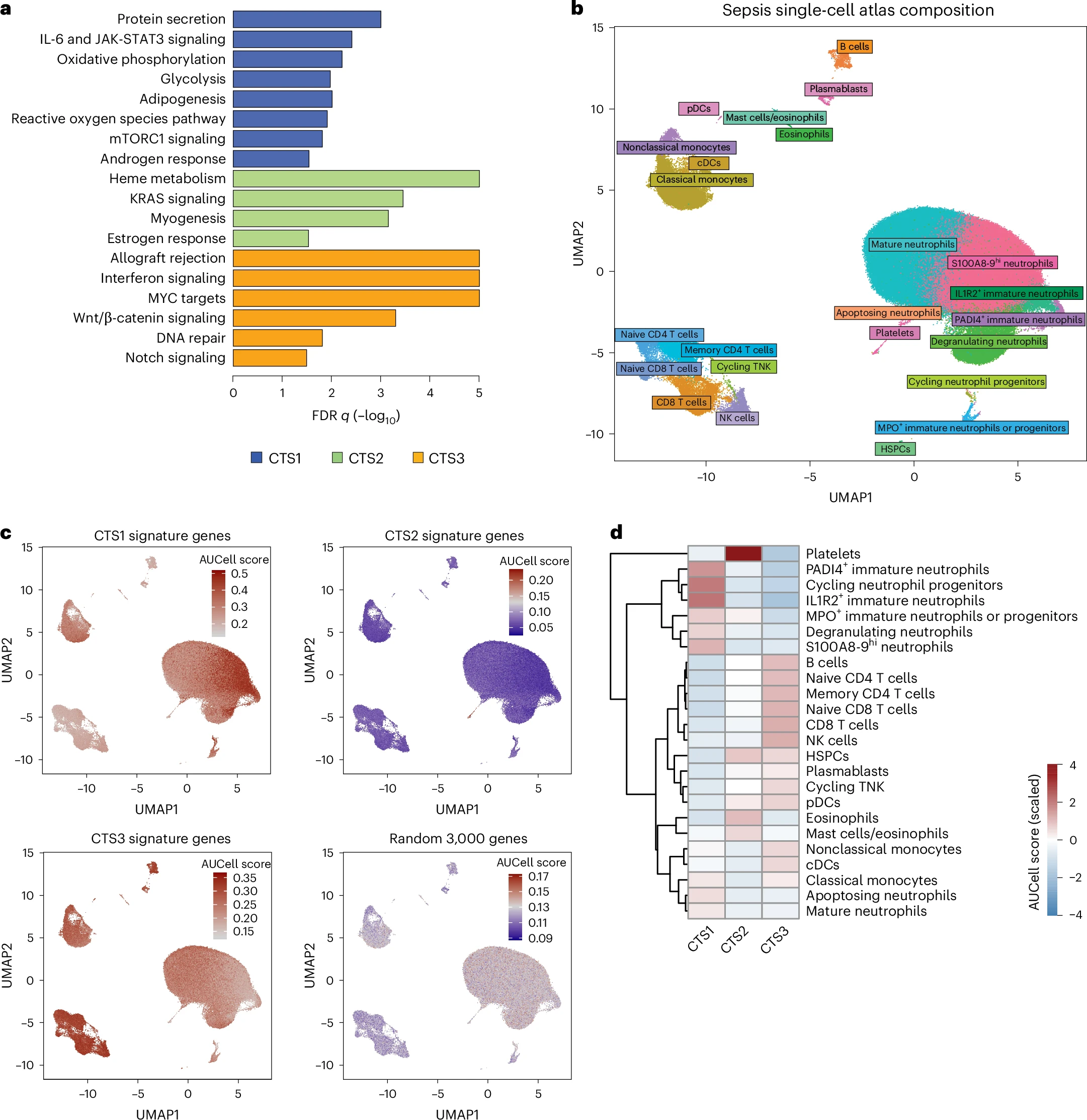
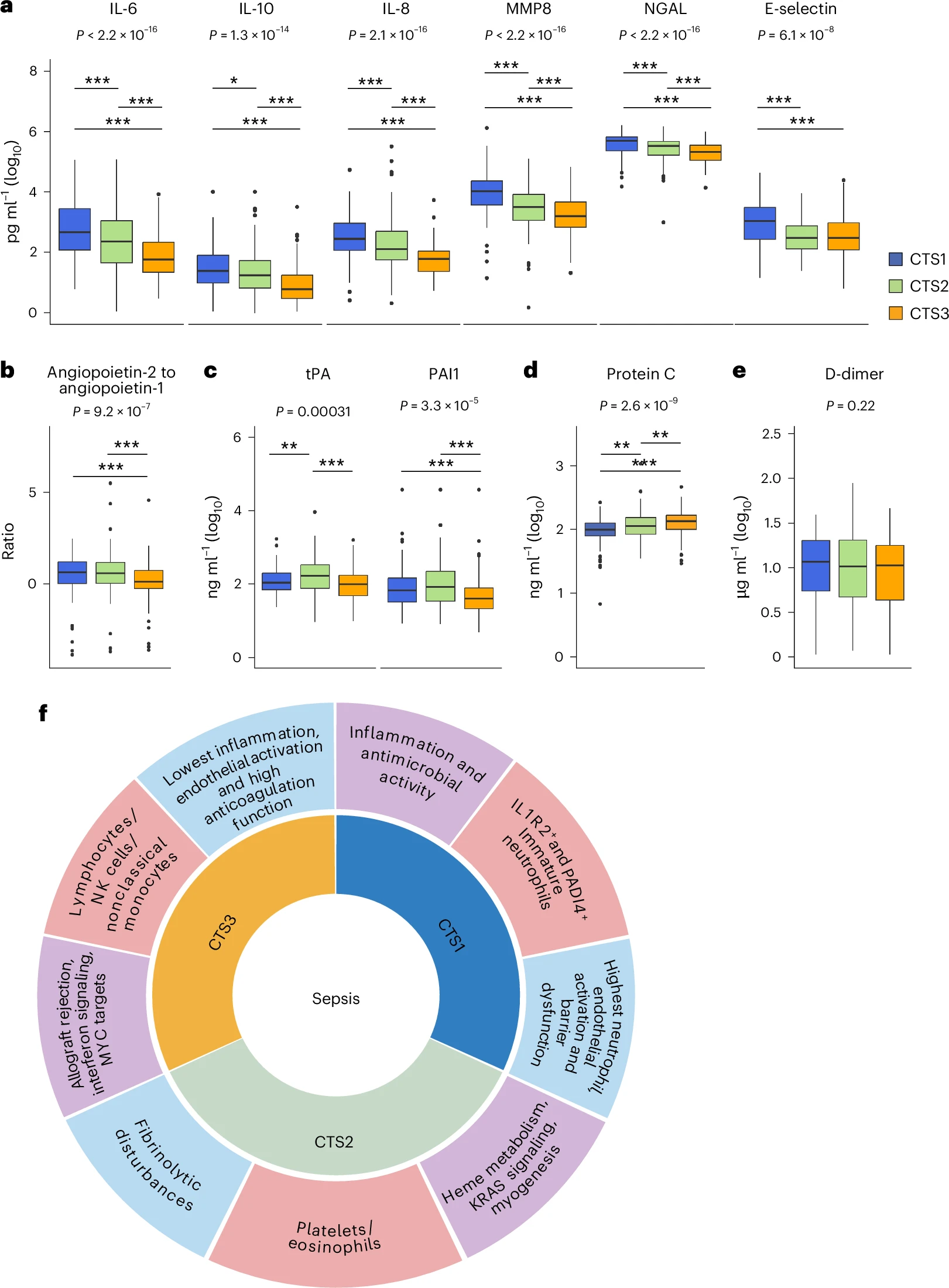
Sepsis is a life-threatening condition driven by a maladaptive host response to infection. To establish a standardized blood transcriptomic subtype model, we aggregated blood transcriptomics data from two major sepsis cohorts: the Molecular Diagnosis and Risk Stratification of Sepsis (MARS) project (n = 678 sampled on intensive care unit admission; ClinicalTrials.gov registration no. NCT01905033) and the Genomic Advances in Sepsis (GAinS) study (n = 444 sampled on intensive care unit admission and n = 817 follow-up samples; ClinicalTrials.gov registration no. NCT00131196). We demonstrate a strong interconnection across three separate classification methods, resulting in the proposed groupings of three consensus transcriptomic subtypes (CTSs). The distinguishing characteristics of CTS1 included gene activation of typical inflammatory pathways, more pronounced endothelial activation and an overall immature neutrophil theme. CTS2 was characterized by gene activation of a heme metabolism pathway, fibrinolytic disturbances and platelet and eosinophil signatures. CTS3 was associated with genes involved in the activation of allograft rejection, interferon signaling and anticoagulation functions, together with lymphocyte and nonclassical monocyte features. Evaluating CTS classification in independent patient cohorts, specifically the vasopressin vs noradrenaline as initial therapy in septic shock (VANISH) randomized controlled trial (n = 176; ISRCTN registration no. ISRCTN20769191) and patients hospitalized with suspected sepsis at a district hospital in Uganda (n = 128), ascertained the robustness of our approach. Notably, post hoc analysis of a pseudo-randomized cohort, along with a reanalysis of the VANISH trial data, unmasked a harmful signal in CTS2-assigned patients treated with corticosteroids. The CTS classification method aligns diverse sepsis transcriptomic subgroupings into a robust, reproducible framework, thereby enabling biological interpretation and potentially assisting aspects of clinical trial design to advance precision medicine in sepsis.